CARLsim4: An Open Source Library for Large Scale, Biologically Detailed Spiking Neural Network Simulation using Heterogeneous Clusters
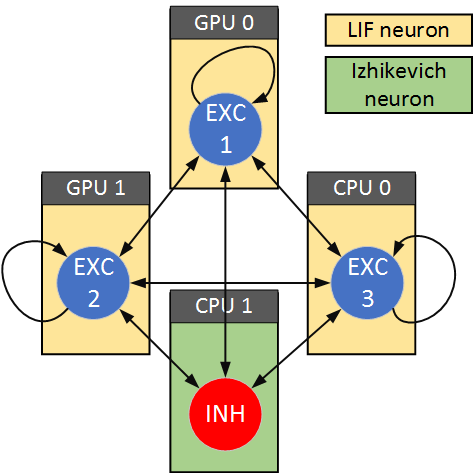
Spiking neural network (SNN) models describe key aspects of neural function in a computationally efficient manner and have been used to construct large-scale brain models. However, large-scale SNNs have proved challenging to implement in practice, due to the required memory to store the network structure and the computational power needed to quickly solve the neuronal dynamics. In addition, it is often necessary to optimize a large number of state variables and open parameters in order to stabilize these large-scale dynamical systems. To meet these challenges, we have developed CARLsim4, a user-friendly, GPU-accelerated SNN library written in C/C++ that is capable of simulating biologically detailed neural models without sacrificing performance. Building on the demonstrated efficiency and scalability of earlier releases, the present release allows for the simulation of up to 10 6.5 neurons and 10 8.5 synapses split over multiple CPU and GPU nodes in a cluster. A benchmark suite is provided to help partitioning a large-scale neural network. In addition, CARLsim4 provides native support for multiple neural models, such as, Leaky-Integrate-and-Fire neuron, 9-parameters Izhikevich neuron, and multi-compartment model. Similar to prior releases, CARLsim4 provides a range of spike-based synaptic plasticity mechanisms and topographic synaptic projections, as well as a network-level parameter tuning interface. CARLsim4 further improves the usability of earlier releases by including multiple platform compatibility (Linux, Mac OS X, and Windows), rigorous code documentation (including an extensive user guide and tutorials), a test suite for functional code verification, and a MATLAB toolbox for the visualization and analysis of neuronal, synaptic, and network information. Benchmarking results demonstrate a 60x speedup for multi-GPU implementations over a single-threaded CPU implementation, making CARLsim4 well-suited for large-scale SNN models in the presence of real-time constraints (e.g., for SNN models of the order of 8.6 million neurons and 0.48 billion synapses using 4 GPUs that interact with neuromorphic sensors or control neurorobotics platforms).
To promote its use among the computational neuroscience and neuromorphic engineering communities, CARLsim4 is provided as an open-source C++ package, which can be freely obtained from https://github.com/UCI-CARL/CARLsim4 and www.socsci.uci.edu/~jkrichma/CARLsim.
Publications
2018
Chou, Ting-Shuo; Kashyap, Hirak J; Xing, Jinwei; Listopad, Stanislav; Rounds, Emily L
International Joint Conference on Neural Networks (IJCNN), 2018.
@conference{choucarlsim,
title = {CARLsim 4: An Open Source Library for Large Scale, Biologically Detailed Spiking Neural Network Simulation using Heterogeneous Clusters},
author = {Ting-Shuo Chou and Hirak J Kashyap and Jinwei Xing and Stanislav Listopad and Emily L Rounds},
url = {http://www.socsci.uci.edu/~jkrichma/Chou-Kashyap-CARLsim4-IJCNN2018.pdf, paper},
year = {2018},
date = {2018-01-01},
booktitle = {International Joint Conference on Neural Networks (IJCNN)},
keywords = {},
pubstate = {published},
tppubtype = {conference}
}
Kashyap, Hirak J; Detorakis, Georgios; Dutt, Nikil; Krichmar, Jeffrey L; Neftci, Emre
A Recurrent Neural Network Based Model of Predictive Smooth Pursuit Eye Movement in Primates Conference
International Joint Conference on Neural Networks (IJCNN), 2018.
@conference{kashyaprecurrent,
title = {A Recurrent Neural Network Based Model of Predictive Smooth Pursuit Eye Movement in Primates},
author = {Hirak J Kashyap and Georgios Detorakis and Nikil Dutt and Jeffrey L Krichmar and Emre Neftci},
url = {http://www.socsci.uci.edu/~jkrichma/Kashyap-PredicitvePursuit-IJCNN2018.pdf, paper},
year = {2018},
date = {2018-01-01},
booktitle = {International Joint Conference on Neural Networks (IJCNN)},
keywords = {},
pubstate = {published},
tppubtype = {conference}
}
2015
Beyeler, Michael; Carlson, Kristofor D; Chou, Ting-Shuo; Dutt, Nikil D; Krichmar, Jeffrey L
CARLsim 3: A user-friendly and highly optimized library for the creation of neurobiologically detailed spiking neural networks Proceedings Article
In: 2015 International Joint Conference on Neural Networks, IJCNN 2015, Killarney, Ireland, July 12-17, 2015, pp. 1–8, 2015.
@inproceedings{DBLP:conf/ijcnn/BeyelerCCDK15,
title = {CARLsim 3: A user-friendly and highly optimized library for the creation of neurobiologically detailed spiking neural networks},
author = {Michael Beyeler and Kristofor D Carlson and Ting-Shuo Chou and Nikil D Dutt and Jeffrey L Krichmar},
url = {https://doi.org/10.1109/IJCNN.2015.7280424},
doi = {10.1109/IJCNN.2015.7280424},
year = {2015},
date = {2015-01-01},
booktitle = {2015 International Joint Conference on Neural Networks, IJCNN 2015,
Killarney, Ireland, July 12-17, 2015},
pages = {1--8},
crossref = {DBLP:conf/ijcnn/2015},
keywords = {},
pubstate = {published},
tppubtype = {inproceedings}
}




